MCB111: Mathematics in Biology (Fall 2024)
week 12:
Genetic Switches
Preliminars
Present all your reasoning, derivations, plots and code as part of the homework. Imagine that you are writing a short paper that anyone in class should to be able to understand. If you are stuck in some point, please describe the issue and how far you got. A jupyter notebook if you are working in Python is not required, but recommended.
A fast and robust oscillator
In class, we discussed the repressilator by Elowitz & Leibler, a synthetic network implemented in E. coli that produces oscillations. The repressilator turned out to be not very robust, as only 40% of the cells presented oscillations. In this homework, we are going to implement a different oscillator that depends on two genes with both negative and positive feedback loops: “A fast, robust and tunable synthetic gene oscillator” by J. Stricker et al.
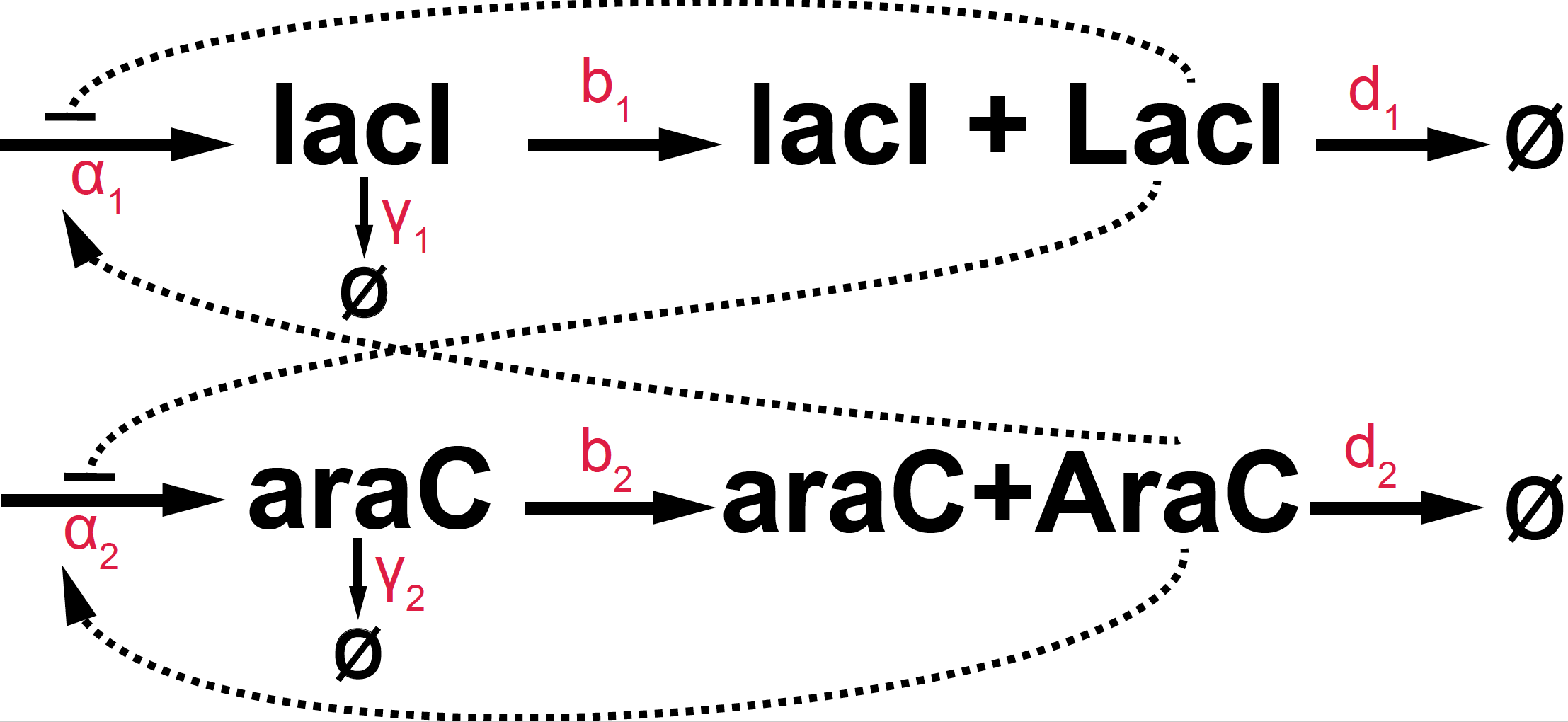
Figure 1. Basic diagram of the Stricker oscillator.
A diagramtic representation of this oscillator is given in Figure 1. In terms of the actual parameters:
-
We know that the LacI protein acts as a dimer, and the AraC protein acts as a tetrad, which determines the Hill coefficients for the feedback loops.
-
We assume enzymatic decay for all RNA and protein species with Michaelis-Menten coefficients provided bellow.
| parameter | Description | value |
|---|---|---|
| \(\alpha_1\) | lacI RNA production | 2.5 |
| \(\alpha_2\) | araC RNA production | 2.5 |
| \(\gamma_1\) | lacI RNA degradation | 0.6 |
| \(\gamma_2\) | araC RNA degradation | 0.8 |
| \(n1\) | cooperativity coefficient of LacI binding | 2 |
| \(n2\) | cooperativity coefficient of AraC binding | 4 |
| \(K^+\) | Hill constant for LacI (-) feedback loop | 2 |
| \(K^-\) | Hill constant for AraC (+) feedback loop | 0.003 |
| \(b_1\) | lacI translation rate | 0.4 |
| \(b_2\) | araC translation rate | 0.1 |
| \(d_1\) | LacI degradation rate | 0.8 |
| \(d_2\) | AraC degradation rate | 0.2 |
| \(K\gamma 1\) | Michaelis constant for lacI RNA degradation | 0.01 |
| \(K\gamma 2\) | Michaelis constant for araC RNA degradation | 0.1 |
| \(Kd1\) | Michaelis constant for LacI degradation | 0.01 |
| \(Kd2\) | Michaelis constant for AraC degradation | 0.1 |
-
Propose the kinetic equations of the system.
-
Solve numerically the differential equations, and plot the concentrations of the four species as a function of time, for the particular values provided in the table above.
-
In your hands, is this oscillator more robust (theoretically) than the repressilator?
-
Do you need the Michaelis-Menten enzymatic degradation for the system to oscillate?
-
Can you identify one parameter important to maintain the oscillatory behavior? What is the range of parameters for which there are oscillations.
-
Which parameter would you use to control the period of the oscillations?
-
If you remove the araC gene, does the lacI system with just one negative feedback loop oscillate? How that is compare to the two-gene system of Stricker?
-
The Stricker et al. implementation is slightly different from this one. You can see the details in their supplemental materials. How does your implementation compare to Stricker’s?