MCB111: Mathematics in Biology (Fall 2024)
week 10:
Molecular Population Dynamics as a Markov Process
Stochastic simulation of transcriptional bursting
Sampling fast from an exponential distribution
In the Gillespie algorithm, dwell times \(\tau\) follow a exponential distribution.
The probability density function (PDF) is
\[PDF(t) = W_R e^{-W_R\ t}.\]In order to sample a dwell time \(\tau\), we used the cumulative probability function (CDF)
\[CDF(\tau) = \int_0^{\tau} W_R\ e^{-W_R\ t}\ dt\]such that for a random number \(0\leq x\leq 1\), the sampled dwell \(\tau\) time is given by
\[x = CDF(\tau).\]The CDF of the exponential distribution has a closed form given by
\[CDF(\tau) = \int_0^{\tau} W_R\ e^{-W_R\ t} = 1 - e^{-W_R\ \tau}.\]Then, the sampled time is given by
\[\tau = -\frac{1}{W_R}\ \log(1-x).\]If \(x\) is a random number in \([0,1]\), then \(1-x\) is also a random number in the same range, then we can take the sampled dwell time \(\tau\) given by
\[\tau = -\frac{1}{W_R}\ \log(x).\]Implementing the Gillespie algorithm
For the stochastic process described in Figure 1, the reactions are given by
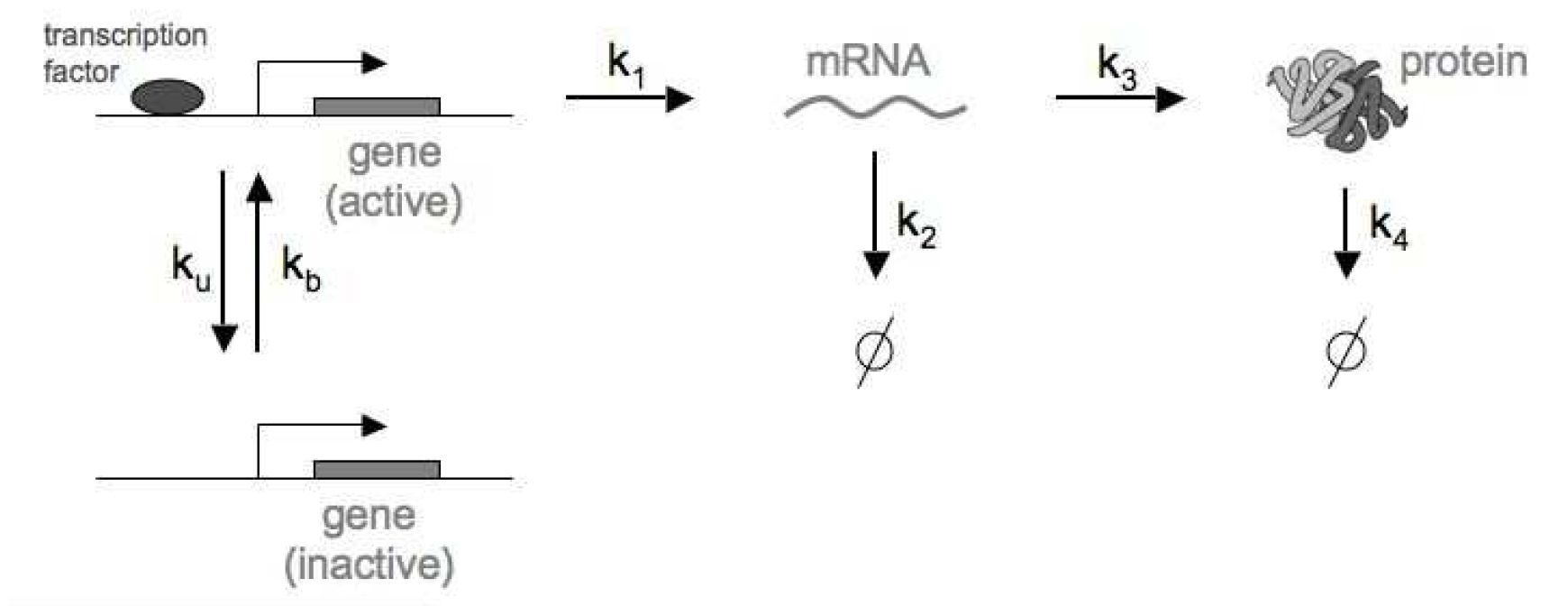
Figure 1. Regulation of a single gene, transcription and translation, as well as RNA and Protein degradation processes.
| r | reaction | propensity \(w_r\) | \(\eta(R)\) | \(\eta(P)\) |
|---|---|---|---|---|
| 1 | \(\mbox{Gene(I)} \overset{k_b}{\longrightarrow} \mbox{Gene(A)}\) | \(k_b\) | \(0\) | \(0\) |
| 2 | \(\mbox{Gene(A)} \overset{k_u}{\longrightarrow} \mbox{Gene(I)}\) | \(k_u\) | \(0\) | \(0\) |
| 3 | \(\mbox{Gene(A)} \overset{k_1}{\longrightarrow} RNA\) | \(k_1\) | \(+1\) | \(0\) |
| 4 | \(RNA \overset{k_2}{\longrightarrow} \emptyset\) | \(k_2 R\) | \(-1\) | \(0\) |
| 5 | \(RNA \overset{k_3}{\longrightarrow} Protein\) | \(k_3 R\) | \(0\) | \(+1\) |
| 6 | \(Protein \overset{k_4}{\longrightarrow} \emptyset\) | \(k_4 P\) | \(0\) | \(-1\) |
Then the Gillespie algorithm works as follows
-
Initialize at \(t = 0\),
\[\begin{aligned} Gact &= 1,\\ R &= 0,\\ P &= 0.\\ \end{aligned}\] -
Keep a current count of \(t\), \(Gact\), \(R\) and \(P\).
- \(Gact = 1\) if the gene is active (A) at time \(t\), or
- \(Gact = 0\) if the gene is inactive (I) at time \(t\).
- \(R\) is the number of RNA molecules at time \(t\).
- \(P\) is the number of protein molecules at time \(t\).
-
Sample a dwell time \(\tau\) from a exponential distribution
\[W_R\ e^{-W_R\ t},\]where the total propensity is given by
\[\begin{aligned} W_R = w_1 + w_4 + w_5 + w_6,\quad &\mbox{if}\, Gact = 0,\\ W_R = w_2 + w_3 + w_4 + w_5 + w_6,\quad &\mbox{if}\, Gact = 1.\\ \end {aligned}\] -
Update the current time \(t\) as
-
Sample one reaction \(r\) according to their propensities
\[P(r) = \frac{w_r}{W_R}.\] -
Update \(Gact\), \(R\) and \(P\)
-
If \(r = 1\), then \(Gact=0 \rightarrow 1\)
-
If \(r = 2\), then \(Gact=1 \rightarrow 0\)
-
If \(r = 3\), then \(R \rightarrow R+1\)
-
If \(r = 4\), then \(R \rightarrow R-1\)
-
If \(r = 5\), then \(P \rightarrow P+1\)
-
If \(r = 6\), then \(P \rightarrow P-1\)
-
-
Update the propensities according to the new values of \(Gact\), \(R\) and \(P\).
-
Go back to sampling a new dwell time, until a maximum time \(T\).